This is the blog for GW students taking Human Evolutionary Genetics. This site is for posting interesting tidbits on: the patterns and processes of human genetic variation;human origins and migration; molecular adaptations to environment, lifestyle and disease; ancient and forensic DNA analyses; and genealogical reconstructions.
GWHEG figure
Tuesday, March 31, 2020
Journal Update 2: Variation in DNA Methylation and Epialleles
In this paper, Zhang et al. use the model plant Arabidopsis to analyze the role of methylation in heterochromatin regulation. Arabidopsis was previously known to show stability in heritability of epialleles across generations. This study found that levels of methylation within euchromatin genes is inversely correlated with heterochromatin methylation. The study also found that genes that are methylated at CG sites at the regulatory regions are more likely to be methylated at non-CG sites and become silenced. The authors suggest that inherent variability in an organism's ability to maintain patterns of DNA methylation can lead to phenotypic changes leading to speciation in Arabidopsis and other plants.
Audrey Tjahjadi
Link to original article: https://www.pnas.org/content/117/9/4874
Epigenetics and Addiction

Addiction, and abuse of addictive substances, fundamentally alters how the brain's reward pathway functions, causing altered behavior and lifestyle defining an "addict". However, the epigenetic mechanisms of how this occurs on a cellular/molecular level are incompletely characterized and understood. A recent paper by Hamilton and Nestler shows that epigenetic modifications, specifically histone modifications, are associated with addictive behavior. They also discuss neuroepigenetic editing as a means to identify the causal mechanisms that drive an addicted state. Altogether this research should help scientist understand what causes and defines an addictive state, as well as potentially providing useful therapies for recovering addicts.
Ryan McRae Potluck 3/31/20
BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses
In the article Wang et al. examine the effects of ten epigenetic factors and their impact on the virus pseudorabies (PRV). They found that the inhibition of the "master regulator of gene expression" BRD4 boosted the immune response against the virus (PRV). This is because BRD4 inhibition caused the DNA damage response by inducing cellular instability. Thus, increasing the hosts resistance to infection because, the inhibitory effect of BRD4 is attributed to the reduction of viral attachment. So if the virus can't attach properly then it cannot infect that cell. Other research on BRD4 suggests that it can be used against cancers such as prostate and breast cancer because BRD4 inhibition decreases RAD51 expression which would damage the cancer DNA cells.
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008429
Pot-luck 3/30/2020
Annelise Beer
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008429
Pot-luck 3/30/2020
Annelise Beer
Monday, March 30, 2020
Epigenome-wide meta-analysis of blood DNA methylation in newborns and children identifies numerous loci related to gestational age
In this study, researchers looked at methylation in the fetal umbilical cord blood, examining the association between methylation and gestational age.
They found that premature babies have different levels of methylation, although most of these differences do not persist into childhood. However, for 17% of these changes, they persist, potentially leading to long-term health effects. Overall 325 genes were identified as having differences in methylation.
-Zac Truesdell, potluck 3/31/2020
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7050134/
They found that premature babies have different levels of methylation, although most of these differences do not persist into childhood. However, for 17% of these changes, they persist, potentially leading to long-term health effects. Overall 325 genes were identified as having differences in methylation.
-Zac Truesdell, potluck 3/31/2020
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7050134/
Epic Genetics - Wheat!
Epic Genetic: the hidden story of wheat
Wheat is one of the main sources of nutrition for human life throughout the world and can inhabit vast geographical areas with varying environments. This study of hundreds of wheat lines worldwide offers insight on wheat’s great ability for local adaptation. Despite wheat’s genome being mapped in previous years, there is much more than studying it’s base pairs that show how wheat is such a widely successful crop. These new findings show that crop evolution is linked with phenotypic changes to agricultural conditions. Not only is this exciting news for current genotyping methods, but it also gives scientists new ways to ensure future crop viability. In the future, breeders may be able to equip wheat with climate resistant armor. Studying the epigenetics of wheat and other staple crops can ensure that farmer’s grow the crop for their local environment.
https://www.earlham.ac.uk/newsroom/epic-genetic-hidden-story-wheat
Kristin Carline | potluck contribution | 3/30/20
Nature Update 1/5: A genomic and epigenomic atlas of prostate cancer in Asian populations
This article posits its research on analyzing whole genome, whole-transcriptome and DNA methylation for tumors represented in prostate cancers from asian populations because most data acquired for prostate cancer is derived from western populations. Yields demonstrate that there is a difference in the distribution of genetic expression of prostate lesions from those that were established by The Cancer Genome Atlas. Chinese patients had genomic alterations that were distinct from Western populations. These patients had 41% of their tumors caused by mutations in FOXA1 and 18% each had deletions in ZNF292 and CHD1. FOXA1 is a pioneer factor that targets the androgen response and has demonstrated its self as having a role in the oncogenesis of prostate cancer. Increase levels of FOXA1 are associated with poor prognosis where decreased level of FOXA1 are predict good prognosis. FOXA1 was the highest mutated gene in Chinese cohorts where as in Western populations it covered the entire coding sequence .
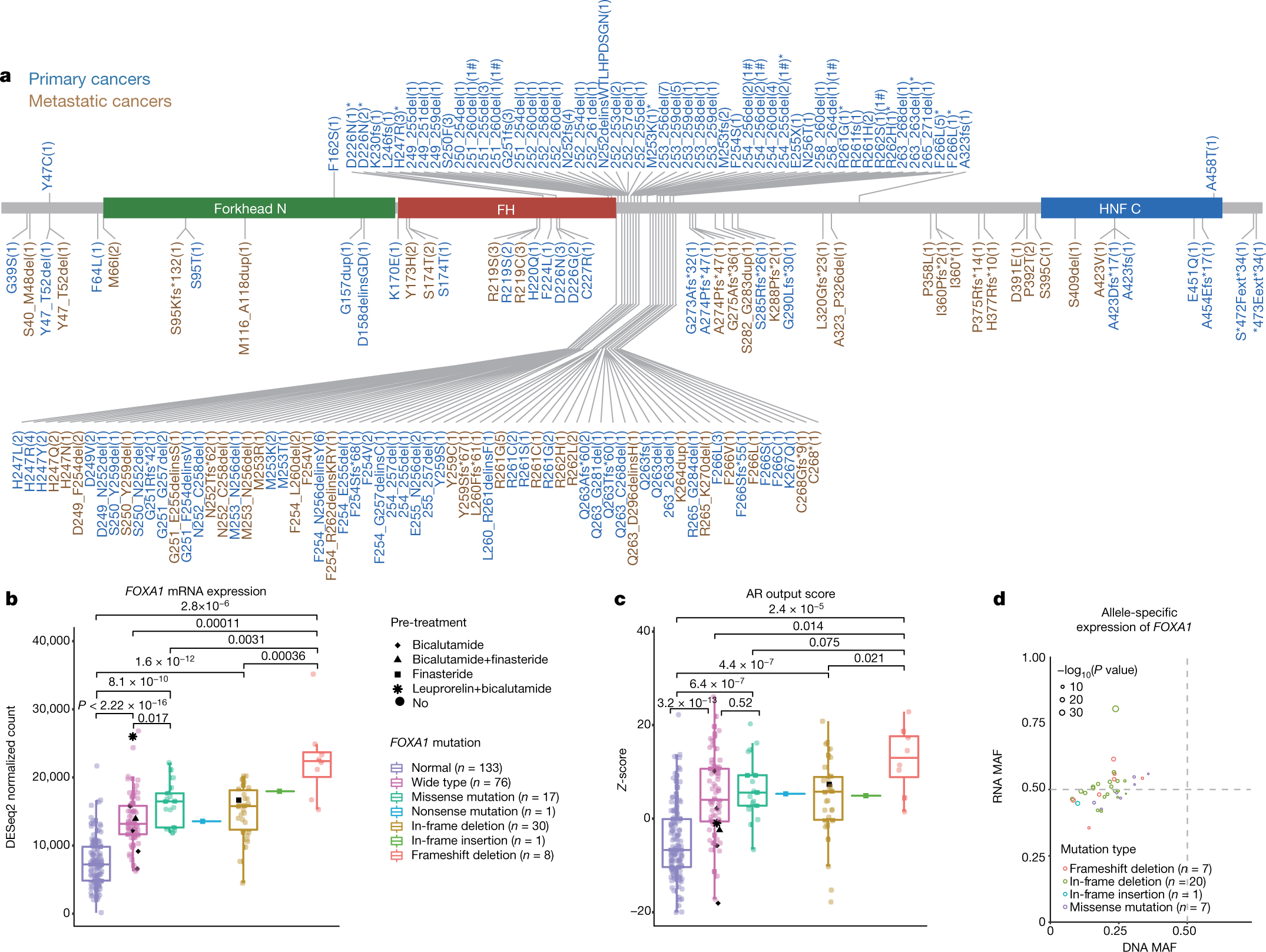
https://www.nature.com/articles/s41586-020-2135-x
Warrenkevin Henderson
Nature Journal Post 1/5
Epigenetic dynamics in infancy and the impact of maternal engagement
A study led by a team at University of Virginia shows that early parental care might influence the development of oxytocin system in infants. Oxytocin is a hormone in the human nervous system that is associated with forming social relationship, care giving and other social behaviours. Through observing 101 mothers and their babies, the team finds that although there is no changes in the level of DNA methylation in the mothers, the babies' saliva samples show some different result.
This study finds that babies who experienced a higher level of involved play with their mothers show a decrease in methylation, which leads to the development of more oxytocin receptors. This result suggests that successful interaction between babies and their care givers at early developmental stage could have an impact on how the oxytocin receptors coding genes express themselves. It could influence the babies' social relationship development later in life.
Monica Cheung– March 30, 2020
Sunday, March 29, 2020
Differential DNA methylation of vocal and facial anatomy genes in modern humans
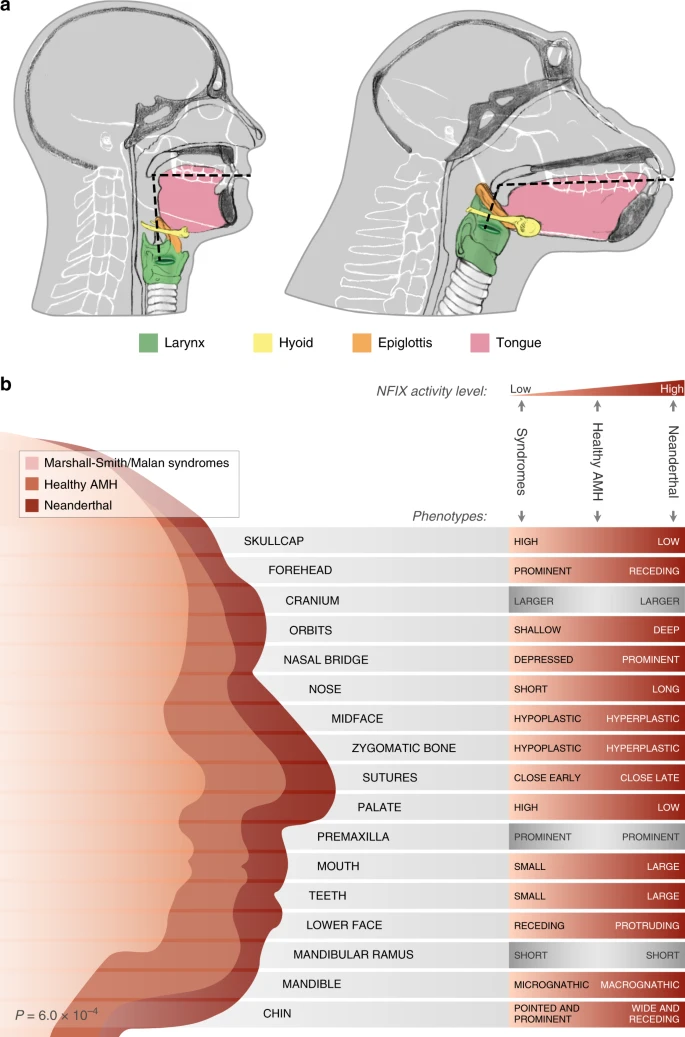 |
| NFIX regulation in AMH, Neanderthals, and Marshall-Smith/Malan Syndromes |
In the past decade, researchers have published significantly more studies on human
evolutionary epigenetic studies. These studies often focus
on DNA methylation, an epigenetic mechanism. One method for determining regulatory
elements affected by methylation is reconstructing and creating experimental methylation
maps. David Gokhman et al. recently published a study on the effects of
DNA methylation of vocal and facial anatomy genes in modern humans. His sample
was comprised of ancient and modern humans as well as six chimpanzees. These samples
produced 69 skeletal DNA methylation maps and identified human-specific methylation
states in 588 genes. Most of these changes are involved in vocal and facial anatomy.
Specifically, Gokhman et al. found extensive hypermethylation in SOX9,
ACAN, COL2A1, NFIX, and XYLT1. They suggest that these methylation changes shaped
the modern human vocal track and face.
Joshua Porter
Potluck – 03/31/2020
Link to paper: https://www.nature.com/articles/s41467-020-15020-6.pdf
Monday, March 23, 2020
Genome‐wide analysis of DNA methylation in endometriosis using Illumina Human Methylation 450 K BeadChips
The aim of this study was to conduct a GWAS investigating variations in DNA methylation in women with endometriosis. Endometriosis is a gynecological disorder that affects approximately 1 in 10 women worldwide, but takes, on average, ten years to get a diagnosis. As of right now, the only way to be diagnosed with endometriosis is surgical excision and biopsy of lesions, a costly and painful method. 12,159 differentially methylated CpG sites and 375 differentially methylated promoter regions were identified; these played roles in inflammatory response, cell adhesion, negative response to apoptosis, and other factors that play a role in the manifestation of endometriosis. It will be interesting to see if epigenetic markers begin to be used more regularly as diagnostic tools as an alternative to surgery in this and other illnesses.
https://onlinelibrary.wiley.com/doi/pdf/10.1002/mrd.23127?casa_token=GVs3q-mtnksAAAAA:1l2YXHVJ_4-Il6IQ-X7Q5xRvEXZQjIRFRNRFyBgZFE3w3NhrpXs9BULZ-QJTay3R2WCP7kHoNuHb1Q
Rachel Nelson
03/23/20
Modified Bugs Boost Bee Health
Because Honey Bees are essential pollinators, a lot of work has focused on their health and productivity. The present article highlights a study conducted by Leonard et al. Previous research has demonstrated that injecting honey bees with double stranded RNA (dsRNA) initiates the RNA interference immune response. This mechanism occurs not only in the host bee itself, but also any parasites inhabiting the bee, and can be used to "knock down" parasite genes that are used for viral replication. There are several drawbacks to this method, including how expensive and prone to degradation dsRNA is, and unintended effects. Leonard et al. was able to make Honey Bees continuously synthesize their own dsRNA, in a much more stable form; this was found to suppress deformed wing virus (WWV), a major threat to this species.
Rachel Nelson
3/23/20
Nature Reviews Genetics Post #2
Sunday, March 22, 2020
Divergent Whole-Genome Methylation Maps of Human and Chimpanzee Brains Reveal Epigenetic Basis of Human Regulatory Evolution
The authors of this paper, Divergent Whole-Genome Methylation Maps of Human and Chimpanzee Brains Reveal Epigenetic Basis of Human Regulatory Evolution, explored the different patterns of DNA methylation in the prefrontal cortex between humans and chimpanzees and how these affect phenotypes. To carry out this investigation, the team employed whole genome sequencing of bisulfite converted DNA. They show that differences in DNA methylation in part explain divergence in gene expression between humans and chimpanzees and that many differentially methylated genes are associated with neurological disorders and cancers.
Link to Paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3511995/
Elaine Miller – Potluck 3/24/2020
Thursday, March 19, 2020
Are twins the key to study epigenetic changes?

Similar to many scientific disciplines, twins present an unparalleled opportunity to study environmental and genetics influences. This has never been more true than epigenetic studies, which intertwine genetics and environment, that having monozygotic twins creates a strong methodological component to study design. This article review the methodology of using twins and demonstrates that the reduced genetic variation in study twins is so beneficial that smaller sample sizes of twins are still much more informative than other research studies with larger participation.
Jack Richardson - Potluck
Link to paper: https://www.sciencedirect.com/science/article/abs/pii/S0149763419306876
Monday, March 16, 2020
The effects of bariatric surgery on DNA methylation Potluck 3/24/20 A. Williams
In this recent study, the researchers use a longitudinal study approach in order to show direct changes in 4857 CpG methylation sites. Among many variables, the researchers looked at methylation before and after 40 patients underwent bariatric surgery and tracked their levels for 12 months afterwards. As their BMI decreased, so did their measure for biological age and epigenetic age acceleration. Of the 4857 CpG sites with methylation changes, 420 were associated with metabolic traits and genes. Two transcription factors were of increased interest due to their correlation with adipocyte development and type 2 diabetes: TCF7L2 and LMO2. The authors stress the clinical importance of these findings and find significant evidence that bariatric surgery will help long term with patients' weight loss and maintenance as well as prevent against metabolic disorders due to the drastic epigenetic changes associated with it.
-Alexis Williams
Fraszczyk et al. Clinical Epigenetics (2020) 12:14
https://doi.org/10.1186/s13148-019-0790-2
https://doi.org/10.1186/s13148-019-0790-2
Photo: https://doi.org/10.1016/j.celrep.2013.03.018
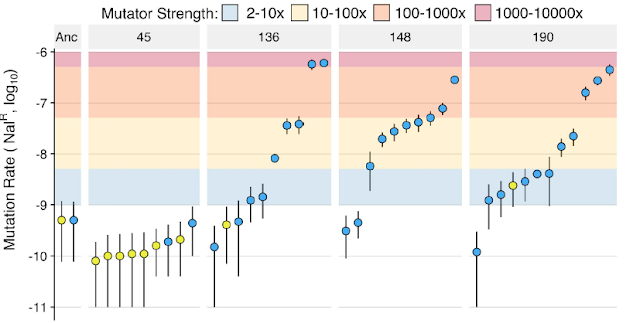
High Mutation Rate Variation in Gut Commensal Bacteria PLOS Bio Update #3 A. Williams
This study measures the mutation load and mutation rate of E. coli in lab mice, a rare paper in observing the evolution of gut commensal bacteria in its natural environment. They find mutations in DNA polymerase III which cause hypermutations, roughly 1000x higher than wild type, that last for over 1000 generations. The rapid mutation rate does produce novel, beneficial mutations that were expected to reach fixation given the fast generation time of the bacteria. However, the researchers found that the beneficial mutations only underwent partial sweeps, not fixation. They conclude that instead of positive selection acting on beneficial mutations, a different type of selection that instead maintains a large range of mutation rate variation (from 4-600 mutations per generation) is acting on this bacteria most strongly. This explains the mechanism for why we see similar community genetic compositions of gut commensal bacteria as this study in human microbiome research as well.
-Alexis Williams
Ramiro RS, Durão P, Bank C, Gordo I (2020) Low mutational load and high mutation rate variation in gut commensal bacteria. PLOS Biology 18(3): e3000617. https://doi.org/10.1371/journal.pbio.3000617
-Alexis Williams
Ramiro RS, Durão P, Bank C, Gordo I (2020) Low mutational load and high mutation rate variation in gut commensal bacteria. PLOS Biology 18(3): e3000617. https://doi.org/10.1371/journal.pbio.3000617
Tuesday, March 10, 2020
Using genetics to discern the paternal ancestry of Brazilian populations - Journal update
This BMC article gives an interesting study linking together historical and genetic data. By looking at the Y-chromosome diversity in the Brazilian population they are able to discern ancestry of male immigrants into the population. They see variation in different areas, reprinting migrants from different areas, which connect well to historical information about colonisation of Brazil. This is a useful way to compare and contrast the maternal and paternal lineages of populations particularly in areas which have a long history of migration.
Jack Richardson - BMC Evolutionary Biology update.
Link to article - https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-020-1579-9
Ethics, Values, and Responsibility: Genome editing from a medical perspective
 The editors of the AMA journal of ethics expressed their views on human genome editing in a letter published in December 2019. Their medical perspective on the issue is intriguing, as they see it is their profession's responsibility to cure disease and alleviate suffering wherever possible. Undoubtedly, genome editing is a great leap forward in medically treating many people. Specifically, they make the distinction between somatic cell editing and germline editing. They state that while somatic cell editing should pose little to no ethical dilemma, it is irresponsible to artificially edit the genome of generations of humans that don't exist, citing potential eugenic uses for the technology. One thing is clear to the authors: medicine has a responsibility to treat people using every tool at their disposal, but not necessarily at the cost of human rights and dignity.
The editors of the AMA journal of ethics expressed their views on human genome editing in a letter published in December 2019. Their medical perspective on the issue is intriguing, as they see it is their profession's responsibility to cure disease and alleviate suffering wherever possible. Undoubtedly, genome editing is a great leap forward in medically treating many people. Specifically, they make the distinction between somatic cell editing and germline editing. They state that while somatic cell editing should pose little to no ethical dilemma, it is irresponsible to artificially edit the genome of generations of humans that don't exist, citing potential eugenic uses for the technology. One thing is clear to the authors: medicine has a responsibility to treat people using every tool at their disposal, but not necessarily at the cost of human rights and dignity.https://journalofethics.ama-assn.org/article/ethics-values-and-responsibility-human-genome-editing/2019-12
Ryan McRae Potluck 3/10/20
Monday, March 9, 2020
CRISPR ethics: moral considerations for applications of a powerful tool
The CRISPR (Clustered Regularly Interspaced Short
Palindromic Repeats)-Cas9 (CRISPR-associated protein 9) system has become a
regular component of genomic engineering studies. However, public support and
understanding of these studies remains uncertain. Recently, Brokowski and Adli
outlined the various ethical issues related to CRISPR and proposed a framework
for regulating future studies. Their review particularly highlighted the uncertainty
associated with human germline genetic modifications and their risks. Another concern
is the likely possibility of class inequality affected access to CRISPR
technology and subsequent price gouging. While human clinical CRISPR trials
will be necessary at some point, these cannot be established without a firm ethical
framework. Overall, they recommend the establishment of an organization, such
as the Association for Responsible Research and Innovation in Genome Editing
(ARRIGE), to help address the ethical considerations of human genome editing.
Joshua Porter - Potluck 03/10/2020
The Ethics of Gene Editing in Animals
Genetically modified animals are a common practice in many fields of science and there is a need to explain the reasoning behind why gene editing in animals is even needed and the ethical explanation for it. This paper reviewed 115 articles to determine their arguments for and against animal gene editing. The most widely described reason was for the applications of modified animals for questions of human health. Other arguments included the efficiency of gene editing for producing organisms that are more beneficial for human life, such as editing disease-carrying mosquitoes. Arguments concerning using gene edited animals to increase animal welfare and improve the environment had both supporters and detractors. Supporters for the animal welfare argument claim that animals could be edited to feel less pain, while others say that it could result in increased numbers of transgenic animals killed for science. For the environmentalist argument, supporters claim that gene editing could be used for as an alternative for insecticides; others claim that gene editing in livestock could increase greenhouse gas production. Overall, the question of why gene editing is necessary is still contentious, even as its use continues to grow.
Audrey Tjahjadi
Link to original article: https://royalsocietypublishing.org/doi/pdf/10.1098/rstb.2018.0106
Audrey Tjahjadi
Link to original article: https://royalsocietypublishing.org/doi/pdf/10.1098/rstb.2018.0106
The CRISPR Zoo
This article addresses using CRISPR to edit genes in animals, and how this could benefit humans or bring species, such as the wooly mammoth, back from extinction. Some examples include editing chicken genes so people are no longer allergic to eggs, sterilizing all female mosquitoes to prevent disease transmission, and editing bee genes so they clean their hives more and are less affected by disease. This raises a lot of ethical questions, such as should humans eat animals that are genetically modified and what are the ecological ramifications of bringing animals back from extinction or sterilizing them?
Rachel Nelson, potluck 2/10/20
https://mon.uvic.cat/tlc/files/2016/10/Welcome-to-the-Crispr-Zoo.pdf
How do you decide what is ethical, and WHO gets to decide?
This news article in science mag, is an interesting perspective from parents considering undergoing gene editing of an embryo to correct a hearing mutation. This work has been presented to them by Denis Rebrikov, who wishes to follow in the footsteps of He Jiankui but this time to correct a hearing mutation, which is present in both the parents. This article gives a different point of view directly from the parents, which creates the discussion, about not only how do you decide what is ethical in terms of gene editing - but also, who gets to decide these guidelines. Are scientists reaching out enough to people that have specific mutation or disorders to ask them about whether their ethical standpoint is and how they would weigh the costs and benefits of these procedures?
Jack Richardson - pot luck
Link to news article:
https://www.sciencemag.org/news/2019/10/deaf-couple-may-edit-embryo-s-dna-correct-hearing-mutation
Photo: https://www.rawpixel.com/image/546019/premium-photo-image-baby-toddler-months-old
Sunday, March 8, 2020
The Human Genome Editing Race: Loosening Regulatory Standards for Commercial Advantage?
Genome editing is becoming a popular tool to create novel gene therapy medicinal products (GTMP). As these technologies are developed, stakeholders in academia, industry and government agencies grapple with testing, approving and regulating these products for use in medicine. The authors of The Human Genome Editing Race: Loosening Regulatory Standards for Commercial Advantage? discuss how the regulatory field must evolve with innovative gene therapy technologies to address questions in how to move forward with clinical testing of GTMPs. The authors point out that each treatment design is different and therefore should be assessed on a case-by-case basis taking into account the quality of preclinical data and the need for the particular treatment. Furthermore, the authors discuss how China and the United States are taking the lead in clinical testing while Europe lags behind. It may be that European researchers are more risk-averse than their Chinese and American counterparts or Chinese and American researchers are hastier in their approach to medical progress.
Scientific Advice Interactions on GTMPs Conducted by the Paul-Ehrlich-Institute between 2013 and the First Half of 2018
Elaine Miller – Potluck 3/10/2020
First use of CRISPR inside a human body -- Potuck 3/10/20, Mackenzie Hepker
Scientists have recently attempted to use CRISPR to cure a genetic disorder called congenital amaurosis, which causes blindness. The mutation causes the loss of function of a protein in the retina that converts light into the signals used by the brain. The researchers conducted retinal surgery on patients with the condition and CRISPR'd out the mutation within the gene, in the hopes that the DNA will ligate and the gene will function normally for the rest of the patients' lives. Results may not be observable for up to a month -- but it's exciting if it works!
https://www.nbcnews.com/health/health-news/doctors-use-crispr-gene-editing-inside-person-s-body-first-n1149711
Mack
Potluck 3/10/2020
https://www.nbcnews.com/health/health-news/doctors-use-crispr-gene-editing-inside-person-s-body-first-n1149711
Mack
Potluck 3/10/2020
Thursday, March 5, 2020
Gene editing strategy for HBB gene mutations -Potluck 3/10/20 A.Williams
Sickle cell disease (SCD) is one of the most highly prevalent diseases in the world with over 300,000 babies born with it each year due to the benefit of heterozygous carriers in preventing malaria. It has recently been the target of CRISPR technology due to the fact that it is caused by a single point mutation on the HBB gene and should therefore be an easier target in a relatively simpler pathway. Over 80% of patients with this disease do not have access to bone transplant treatment. This study announces the creation of two guide RNAs (gRNAs) and their success in targeting the point mutation in human cells. Moreover, they show proof of concept that longer gRNAs are more effective at targeting specific sequences with a lower chance of non-target disruption. Although much more testing is needed before this method is used directly in humans, it is a promising start in creating another treatment option for patients with the most severe form of SCD (HbSS) when bone transplant is not an option.
-Alexis W.
https://doi.org/10.1016/j.gene.2020.144398
-Alexis W.
https://doi.org/10.1016/j.gene.2020.144398
NRF2 negatively regulates primary ciliogenesis and Hh signaling -Plos Biology Update #2 A. Williams
The hyperactivation of the NRF2, a transcription factor involved in protein homeostasis signaling, pathway is known to cause tumor progression through rapid proliferation of cells and increased survivability of these cells under harsh conditions. It is also known for being resistant to both chemotherapy and radiotherapy in cancer patients. Therefore, understanding the mechanisms behind its networks is key in treating patients. The researchers found that not only was NRF2 negatively involved in primary ciliogenesis, the building of organelles involved in extracellular cell signaling through increased expression of p26/SQSTM1 (an autophagy), but also up regulates PTCH1, a negative regulator of Hh signaling which controls development, cell fate, and cell renewal. Critically, the researchers discovered that the destruction of p26 and PTCH1 together was sufficient in stopping NRF2's effects on tumor progression and could be a viable way of treating patients in the future.
-A. Williams
https://doi.org/10.1371/journal.pbio.3000620
Wednesday, March 4, 2020
Neanderthal ‘minibrains’ grown in dish
Neanderthal ‘minibrains’ grown in dish

Scientist are growing "minibrains" to study the influence of ancient DNA by engineering stem cells to include Neanderthal genes. The researchers involved are trying to "recreate" the Neanderthal mind. These organoids are not full, functioning brains, but are a step towards learning more about early brain structure and development. Muotri's group is focusing on a gene we discussed last week (briefly), NOVA1. It plays a role in early brain development and is linked to autism and schizophrenia. Pääbo's research group is also working on organoids and stresses that the organoids are an early developing research method, one that his lab group is also working on, and that the technique can produce mutations. It will be interesting to see how the organoid "minibrain" research develops and whether there will be viable functionally relevant information.
Kailie Batsche
Potluck for 10 March 2020
Monday, March 2, 2020
Researchers map genetic ‘switches’ behind human brain evolution
In this study, scientists mapped regulatory regions in human neurogenesis. I'll be honest, I don't entirely understand what is going on in this paper, but it seems they are looking at chromatin accessibility to determine which genomic regions are active during neurogenesis.
The researchers say one of their most significant findings is a human-specific FGFR2 enhancer which plays a function in human cortical expansion. In addition, by understanding the pathways that are present in neurogensis, this can help us understand which diseases occur when one of these pathways is disrupted.
The press release gives a succinct summary:
https://newsroom.ucla.edu/releases/ucla-map-genetic-switches-behind-human-brain-evolution
The paper:
https://www.sciencedirect.com/science/article/pii/S0092867417314940
Zac Truesdell, potluck, 3/3/2020
The researchers say one of their most significant findings is a human-specific FGFR2 enhancer which plays a function in human cortical expansion. In addition, by understanding the pathways that are present in neurogensis, this can help us understand which diseases occur when one of these pathways is disrupted.
The press release gives a succinct summary:
https://newsroom.ucla.edu/releases/ucla-map-genetic-switches-behind-human-brain-evolution
The paper:
https://www.sciencedirect.com/science/article/pii/S0092867417314940
Zac Truesdell, potluck, 3/3/2020
Evolutionary divergence of neuroanatomical organization and related genes in chimpanzees and bonobos
This paper from our very own CASHP examines brain difference in chimpanzees and bonobos from both an anatomical and genomic level. They find that bonobos brains have reduced in a number of parameters, brain size in both males and females and white matter, and chimpanzees brains have increased. Despite these anatomical changes They see no significant difference in genomic structure, apart from some small truncations in some genes, however no observable difference in protein evolution. Suggestion that bonobos brains have shrunk because of self-domestication, similar to what is hypothesised in humans are discussed.
Jack Richardson - 2-Mar-20
Link to paper: https://www.med.upenn.edu/ngg/assets/user-content/documents/ngg-documents/inpress.bonobo.chimpanzee.neuro.evolutionary.divergence.cortex.pdf
Mapping genetic influences on human brain structure
This paper attempts to create mind maps to measure the way that genes are impacted by the shape, size, and complexity of a brain. By doing this they would be able to pinpoint genes as well as shapes that are more susceptible to disease. To do this they have to use a population-based sample which can also shed light on familial relations and brain disorders. These mind maps would allow scientists to visualize a diseased population/ one with known genetic risk, and individual variations can be mapped onto. Parts of the brain such as grey matter, gyral patterning, and brain asymmetry; also varies with age, gender, handedness, and hemispheric dominance that a map of these features would reveal patterns not observable at the individual level.
Potluck Post
3/3/2020
Annelise Beer
Gene associated with autism also controls growth of the embryonic brain

Although autism spectrum disorder has a strong genetic basis, scientists today have yet to fully understand the interactions that lead to the disorder. A UCLA- led study on Foxp1, gene associated with autism, might provide new insights to the development of autism as well as other intellectual and language impairments. Previous researches on Foxp1 focus mainly on its expression in neurons that are already formed. In this new study, scientists discover that the gene becomes active in embryonic brain far earlier than previously thought. Foxp1 appears to have an intimate effect on "the ability of neural stem cells to replicate and form certain neurons in a specific sequence", according to one of the authors of the paper. This new discovery help us further investigate the potential causes and mechanism of autism.
Gene associated with autism also controls growth of the embryonic brain
https://newsroom.ucla.edu/releases/foxp1-gene-autism-growth-embryonic-brain
Monica Cheung– March 2, 2020
Gene associated with autism also controls growth of the embryonic brain
https://newsroom.ucla.edu/releases/foxp1-gene-autism-growth-embryonic-brain
Monica Cheung– March 2, 2020
Human-like Brain Development in Transgenic Rhesus Monkeys
MCPH1 is a gene encoding a multi-functional protein involved in DNA repair, regulation of cell cycling and apoptosis, and regulation of neurogenesis. This gene has shown to be rapidly evolving and has accumulated seven human-specific amino acid changes and shown differences in levels of expression between humans and rhesus monkeys. 11 transgenic rhesus monkeys with human copies of MCPH1 were produced. These monkeys exhibited more human-like neotenic brain development in the form of slower growth of grey matter volume (164 day delay to reach peak compared to WT). Analysis of a marker gene for brain cell maturation, NeuN, showed transgenic monkeys with significantly lower counts of NeuN presenting cells compared to control. Transcriptomic analyses further showed that transgenic monkeys had higher expression of MCPH1 and downregulation of genes involved in neuronal differentiation and development. Interestingly, the brains of the transgenic monkeys were not found to be enlarged, despite the developmental delays. Thus, this study demonstrates the potential usefulness of using primate models to answer questions of human evolutionary development and also brings up questions about ethical animal experimentation.
Audrey Tjahjadi
Link to original article: https://academic.oup.com/nsr/article/6/3/480/5420749
Hox Timing Determines Limb Placement
Hox Timing Determines Limb Placement

Hox genes are famous for controlling vertebral identity. This study has found that differential timing of Hox genes in the lateral plate mesoderm determines limb placement also. Young et al. explore the placement of limbs in tetrapods and avian species to better understand these concepts. They determined that our earliest ancestors in the Devonian period must have possessed Hox genes due to the development of appendages; however, this opens a whole conundrum of what controls the spatiotemporal pattern of these Hox genes and how the tissues coordinate in the gastrulating embryo so that the forelimb always forms in the correct location. The authors suggest more work with the Hox genes of zebra finches, chickens, and ostriches may show some interesting prospects with retinoic acid expression.
https://www.sciencedirect.com/science/article/pii/S0960982218315987
Kristin Carline | Potluck Contribution | 3/2/20
The influence of AVPR1A genotype on individual differences in behaviors during a mirror self-recognition task in chimpanzees (Pan troglodytes)
This article investigated whether variations in the AVPR1A gene resulted in different mirror self recognition (MSR) behaviors in chimpanzees. This gene has been under examination for quite some time, as it seems to be related to the evolution of complex social behaviors and personality. Males with the DupB+/- gene were more likely to exhibit scratching and agonistic behaviors during the mirror-recognition task, and the authors concluded this is partially explained by the AVPR1A polymorphism. Are we convinced that variations in one gene can result in different behaviors and personality types?
https://onlinelibrary.wiley.com/doi/pdf/10.1111/gbb.12291
Rachel Nelson, March 3rd Potluck
Subscribe to:
Posts (Atom)